
- #Pymol tutorial carlsson lab how to#
- #Pymol tutorial carlsson lab full#
- #Pymol tutorial carlsson lab code#
Machine learning approaches are employed to build models for predicting active compounds (T7). Furthermore, measures for compound similarity are introduced and applied for VS of kinase inhibitor gefitinib (T4) as well as for compound clustering (T5), including the use of maximum common substructures (T6).
#Pymol tutorial carlsson lab how to#
The talktorial topics include how to acquire data from ChEMBL (T1), filter compounds for drug-likeness (T2), and identify unwanted substructures (T3). A conda yml file is provided to ensure an easy and quick setup of an environment containing all required packages. Furthermore, the user is instructed how to work with conda, a widely used package, dependency and environment management tool. Additionally, basic Python computing libraries employed include numpy and pandas (high-performance data structures and analysis), scikit-learn (machine learning), as well as matplotlib and seaborn (plotting). Open source libraries utilized are RDKit (cheminformatics), the ChEMBL webresource client and PyPDB (ChEMBL and PDB application programming interface access), BioPandas (loading and manipulating molecular structures), and PyMOL (structural data visualization).
Open data resources employed are the ChEMBL and PDB databases for compound and protein structure data acquisition, respectively. They start with a topic motivation and learning goals, continue with the main part composed of theoretical background and practical code, and end with a short discussion and quiz, see Fig. in student CADD seminars (talk + tutorial = talktorial). Talktorials are offered as interactive Jupyter notebooks that can be used as tutorials but also for oral presentations, e.g. TeachOpenCADD currently consists of ten talktorials covering central topics in CADD, see Fig.
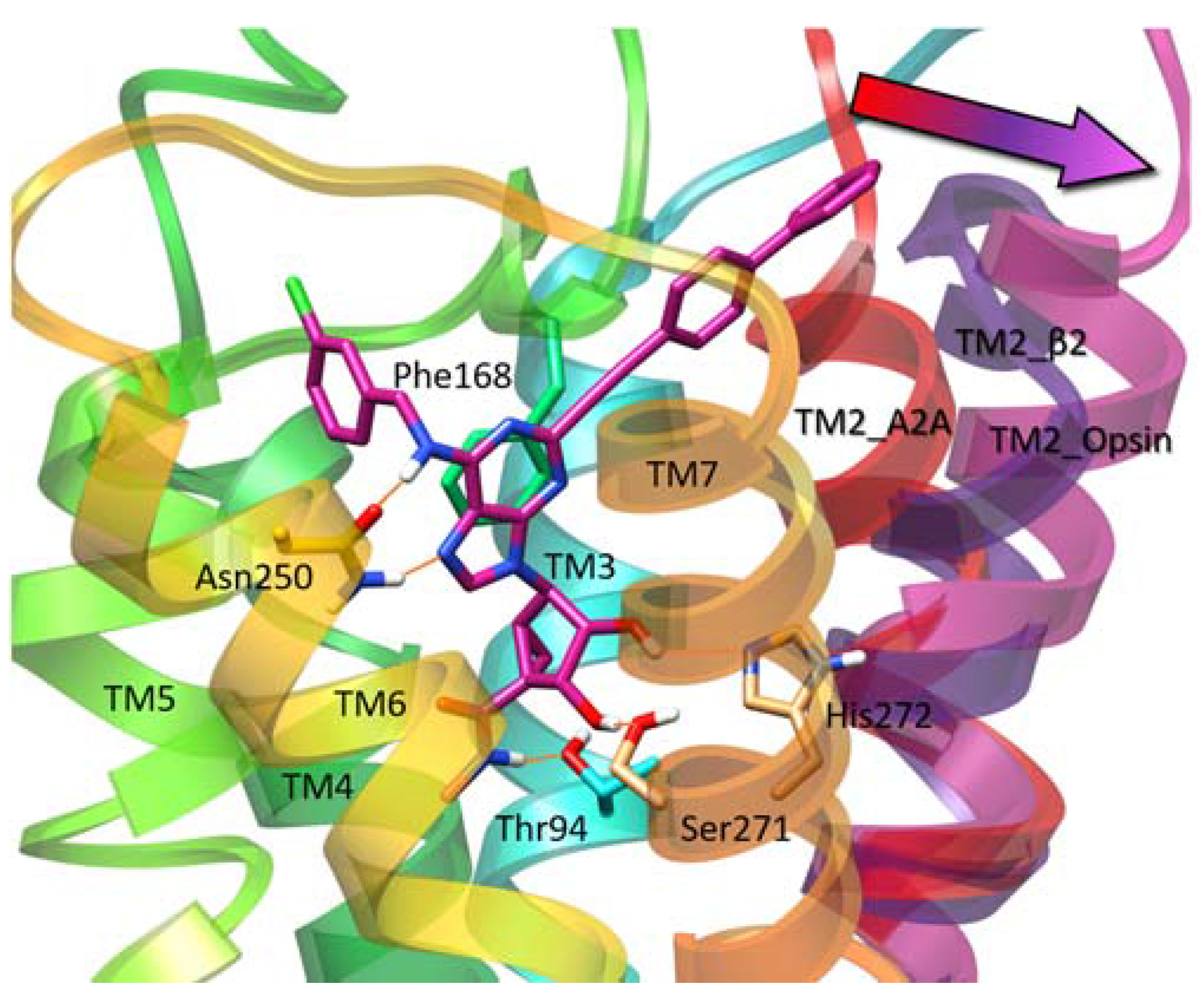
TeachOpenCADD is publicly available on GitHub and open to contributions from the community: (current release: ). Topics build upon one another in the form of a pipeline, which is illustrated at the example of the epidermal growth factor receptor (EGFR) kinase, but can easily be adapted to other query proteins. Interactive Jupyter notebooks are presented for central topics, integrating detailed theoretical background and well-documented practical code. TeachOpenCADD is a novel teaching platform developed by students for students, using open source data and Python packages to tackle various common tasks in cheminformatics and structural bioinformatics. Ĭomplementing these resources, we developed the TeachOpenCADD platform to provide students and researchers new to CADD and/or programming with step-by-step tutorials suitable for self-study training as well as classroom lessons, covering both ligand- and structure-based approaches. On the other hand, examples for educational coding tutorials are the Java-based Chemistry Development Kit (CDK) and the Teach–Discover–Treat (TDT) initiative, which launched challenges to develop tutorials, such as a Python-based virtual screening (VS) workflow to identify malaria drugs. Available examples include the following: On the one hand, graphical user interface (GUI) based tutorials teach CADD basics, such as the web-based educational Drug Design Workshop. While documentation for open access resources is available, freely accessible teaching platforms for concepts and applications in CADD are rare.

#Pymol tutorial carlsson lab full#
, or presented in the form of the web-based search tool Click2Drug, aiming to cover the full CADD pipeline. Comprehensive lists of open resources are reviewed by Pirhadi et al. This combination facilitates and promotes the generation of modular, reproducible, and easy-to-share pipelines for computer-aided drug design (CADD).

#Pymol tutorial carlsson lab code#
Open access resources for cheminformatics and structural bioinformatics as well as public platforms for code deposition such as GitHub are increasingly used in research.


 0 kommentar(er)
0 kommentar(er)
